Note
Click here to download the full example code
Compare topographies¶
This example shows how to compare EEG topographies, based on the method described by McCarthy & Wood 1.
# sphinx_gallery_thumbnail_number = 4
from eelbrain import *
Simulated data¶
Generate a simulated dataset (as in the Cluster-based permutation t-test example)
dss = []
for subject in range(10):
# generate data for one subject
ds = datasets.simulate_erp(seed=subject)
# average across trials to get condition means
ds_agg = ds.aggregate('cloze_cat')
# add the subject name as variable
ds_agg[:, 'subject'] = f'S{subject:02}'
dss.append(ds_agg)
ds = combine(dss)
# make subject a random factor (to treat it as random effect for ANOVA)
ds['subject'].random = True
# Re-reference the EEG data (i.e., subtract the mean of the two mastoid channels):
ds['eeg'] -= ds['eeg'].mean(sensor=['M1', 'M2'])
print(ds.head())
Out:
n cloze cloze_cat n_chars subject
--------------------------------------------
40 0.88051 high 4.125 S00
40 0.17241 low 3.825 S00
40 0.89466 high 4 S01
40 0.13778 low 4.575 S01
40 0.90215 high 4.275 S02
40 0.12206 low 3.9 S02
40 0.88503 high 4 S03
40 0.14273 low 4.175 S03
40 0.90499 high 4.1 S04
40 0.15732 low 3.5 S04
--------------------------------------------
NDVars: eeg
The simulated data in the two conditions:
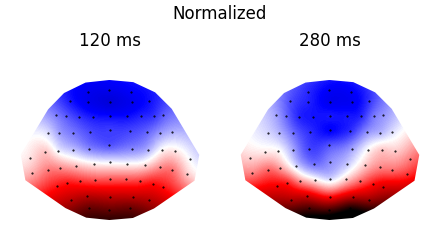
p = plot.TopoArray('eeg', 'cloze_cat', ds=ds, axh=2, axw=10, t=[0.120, 0.200, 0.280])
Test between conditions¶
Test whether the 120 ms topography differs between the two cloze conditions. The dataset already includes one row per cell (i.e., per cloze condition and subject). Consequently, we can just index the topography at the desired time point:
topography = ds['eeg'].sub(time=0.120)
# normalize the data in accordance with McCarth & Wood (1985)
topography = normalize_in_cells(topography, 'sensor', 'cloze_cat', ds)
# "melt" the topography NDVar to turn the sensor dimension into a Factor
ds_topography = table.melt_ndvar(topography, 'sensor', ds=ds)
# Note EEG is a single column, and the last column indicates the sensor
ds_topography.head()
Out:
n cloze cloze_cat n_chars subject eeg sensor
----------------------------------------------------------------
40 0.88051 high 4.125 S00 -0.55174 Fp1
40 0.17241 low 3.825 S00 -0.94975 Fp1
40 0.89466 high 4 S01 -1.0303 Fp1
40 0.13778 low 4.575 S01 -1.5701 Fp1
40 0.90215 high 4.275 S02 -1.3857 Fp1
40 0.12206 low 3.9 S02 -1.5753 Fp1
40 0.88503 high 4 S03 -1.1537 Fp1
40 0.14273 low 4.175 S03 -1.6285 Fp1
40 0.90499 high 4.1 S04 -1.9244 Fp1
40 0.15732 low 3.5 S04 -1.5299 Fp1
ANOVA to test whether the effect of cloze_cat differs between sensors:
test.ANOVA('eeg', 'cloze_cat * sensor * subject', ds=ds_topography)
Out:
<ANOVA: eeg ~ cloze_cat + sensor + cloze_cat x sensor + subject + cloze_cat x subject + sensor x subject + cloze_cat x sensor x subject
SS df MS MS(denom) df(denom) F p
----------------------------------------------------------------------------------------
cloze_cat -0.00 1 -0.00 3.64 9 -0.00 1.000
sensor 1295.07 64 20.24 0.08 576 255.94*** < .001
cloze_cat x sensor 4.93 64 0.08 0.07 576 1.06 .366
----------------------------------------------------------------------------------------
Total 1468.53 1299
>
The non-significant interaction suggests that the effect of cloze_cat does not differ between sensors, i.e., the topographies do not differ, which is consistent with being generated by the same underlying neural sources.
Test two time points¶
Since we’re not interested in condition here, we first average across conditions, i.e., with the goal of having one row per subject:
ds_average = ds.aggregate('subject', drop_bad=True)
print(ds_average)
Out:
n cloze n_chars subject
--------------------------------
40 0.52646 3.975 S00
40 0.51622 4.2875 S01
40 0.51211 4.0875 S02
40 0.51388 4.0875 S03
40 0.53115 3.8 S04
40 0.52163 4.0375 S05
40 0.53789 4.2 S06
40 0.52491 4.1125 S07
40 0.52464 4.15 S08
40 0.52559 3.8875 S09
--------------------------------
NDVars: eeg
In order to compare two time points, we need to construct a new dataset with time point as Factor:
dss = []
for time in [0.120, 0.280]:
ds_time = ds_average['subject',] # A new dataset with the 'subject' variable only
ds_time['eeg'] = ds_average['eeg'].sub(time=time)
ds_time[:, 'time'] = f'{time*1000:.0f} ms'
dss.append(ds_time)
ds_times = combine(dss)
ds_times.summary()
Out:
Key Type Values
------------------------------------------------------------------------------------------------
subject Factor S00:2, S01:2, S02:2, S03:2, S04:2, S05:2, S06:2, S07:2, S08:2, S09:2 (random)
eeg NDVar 65 sensor; -3.62446e-06 - 3.90635e-06
time Factor 120 ms:10, 280 ms:10
------------------------------------------------------------------------------------------------
None: 20 cases
Then, normalize the data in accordance with McCarth & Wood (1985)
topography = normalize_in_cells('eeg', 'sensor', 'time', ds=ds_times)
# "melt" the topography NDVar to turn the sensor dimension into a Factor
ds_topography = table.melt_ndvar(topography, 'sensor', ds=ds_times)
# Note EEG is a single column, and the last column indicates the sensor
ds_topography.head()
Out:
subject time eeg sensor
------------------------------------
S00 120 ms -0.74756 Fp1
S01 120 ms -1.2964 Fp1
S02 120 ms -1.4811 Fp1
S03 120 ms -1.3882 Fp1
S04 120 ms -1.735 Fp1
S05 120 ms -1.3842 Fp1
S06 120 ms -1.2591 Fp1
S07 120 ms -1.3146 Fp1
S08 120 ms -1.4126 Fp1
S09 120 ms -1.0279 Fp1
Plot the topographies before and after normalization:
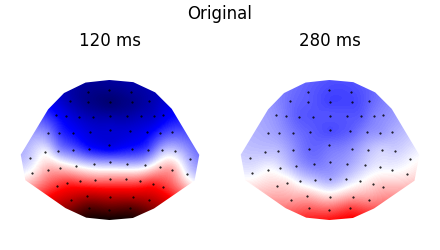
p = plot.Topomap('eeg', 'time', ds=ds_times, ncol=2, title="Original")
p = plot.Topomap(topography, 'time', ds=ds_times, ncol=2, title="Normalized")
Compre the topographies with the ANOVA – test whether the effect of time differs between sensors:
test.ANOVA('eeg', 'time * sensor * subject', ds=ds_topography)
Out:
<ANOVA: eeg ~ time + sensor + time x sensor + subject + time x subject + sensor x subject + time x sensor x subject
SS df MS MS(denom) df(denom) F p
-----------------------------------------------------------------------------------
time 0.00 1 0.00 12.55 9
sensor 1269.88 64 19.84 0.19 576 105.37*** < .001
time x sensor 30.12 64 0.47 0.18 576 2.57*** < .001
-----------------------------------------------------------------------------------
Total 1754.93 1299
>
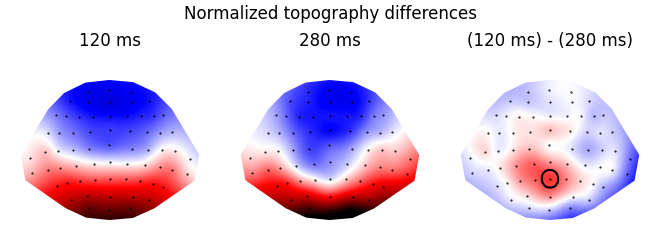
Visualize the difference
res = testnd.TTestRelated(topography, 'time', match='subject', ds=ds_times)
p = plot.Topomap(res, ncol=3, title="Normalized topography differences")
References¶
- 1
McCarthy, G., & Wood, C. C. (1985). Scalp Distributions of Event-Related Potentials—An Ambiguity Associated with Analysis of Variance Models. Electroencephalography and Clinical Neurophysiology, 61, S226–S227. 10.1016/0013-4694(85)90858-2
Total running time of the script: ( 0 minutes 13.670 seconds)