Note
Go to the end to download the full example code
Introduction
Data are represented with there primary data-objects:
Factorfor categorial variablesVarfor scalar variablesNDVarfor multidimensional data (e.g. a variable measured at different time points)
Multiple variables belonging to the same dataset can be grouped in a
Dataset object.
Factor
A Factor is a container for one-dimensional, categorial data: Each
case is described by a string label. The most obvious way to initialize a
Factor is a list of strings:
Factor(['a', 'a', 'a', 'a', 'b', 'b', 'b', 'b'], name='A')
Since Factor initialization simply iterates over the given data, the same Factor could be initialized with:
Factor(['a', 'a', 'a', 'a', 'b', 'b', 'b', 'b'], name='A')
There are other shortcuts to initialize factors (see also
the Factor class documentation):
Factor(['a', 'a', 'a', 'a', 'b', 'b', 'b', 'b', 'c', 'c', 'c', 'c'], name='A')
Indexing works like for arrays:
a[0]
'a'
a[0:6]
Factor(['a', 'a', 'a', 'a', 'b', 'b'], name='A')
All values present in a Factor are accessible in its
Factor.cells attribute:
a.cells
('a', 'b', 'c')
Based on the Factor’s cell values, boolean indexes can be generated:
a == 'a'
array([ True, True, True, True, False, False, False, False, False,
False, False, False])
a.isany('a', 'b')
array([ True, True, True, True, True, True, True, True, False,
False, False, False])
a.isnot('a', 'b')
array([False, False, False, False, False, False, False, False, True,
True, True, True])
Interaction effects can be constructed from multiple factors with the %
operator:
Factor(['d', 'd', 'e', 'e', 'd', 'd', 'e', 'e', 'd', 'd', 'e', 'e'], name='B')
A % B
Interaction effects are in many ways interchangeable with factors in places where a categorial model is required:
(('a', 'd'), ('a', 'e'), ('b', 'd'), ('b', 'e'), ('c', 'd'), ('c', 'e'))
i == ('a', 'd')
array([ True, True, False, False, False, False, False, False, False,
False, False, False])
Var
The Var class is a container for one-dimensional
numpy.ndarray:
Var([1, 2, 3, 4, 5, 6])
Indexing works as for factors
y[5]
6
y[2:]
Var([3, 4, 5, 6])
Many array operations can be performed on the object directly
y + 1
Var([2, 3, 4, 5, 6, 7])
For any more complex operations the corresponding numpy.ndarray
can be retrieved in the Var.x attribute:
array([1, 2, 3, 4, 5, 6])
Note
The Var.x attribute is not intended to be replaced; rather, a new
Var object should be created for a new array.
NDVar
NDVar objects are containers for multidimensional data, and manage the
description of the dimensions along with the data. NDVar objects are
usually constructed automatically by an importer function (see
File I/O), for example by importing data from MNE-Python through
load.mne.
Here we use data from a simulated EEG experiment as example:
<NDVar 'eeg': 80 case, 140 time, 65 sensor>
This representation shows that eeg contains 80 trials of data (cases),
with 140 time points and 35 EEG sensors.
The object provides access to the underlying array…
array([[[-1.10225035e-07, -1.21398829e-06, 1.35355502e-06, ...,
-6.12893745e-07, -2.51649218e-06, -1.33879697e-06],
[ 1.03057616e-06, -1.97609123e-06, 5.55910753e-08, ...,
-1.75102062e-06, -6.06858438e-06, -1.48262419e-06],
[ 2.26162309e-06, -2.76452386e-06, -1.64393195e-06, ...,
-2.56737373e-06, -3.39503569e-06, -1.77351586e-06],
...,
[ 1.27928669e-06, -2.54650493e-06, -1.43902791e-06, ...,
-1.14332247e-06, -7.59471495e-07, -1.39821309e-06],
[ 5.83282737e-07, -2.50177953e-06, -1.96916415e-06, ...,
-1.70881189e-07, -1.97792491e-06, 7.97185353e-07],
[ 1.00898892e-06, -2.53987234e-06, -6.87119668e-07, ...,
1.88518585e-06, -2.40118283e-06, 2.64689298e-06]],
[[ 7.91017764e-07, 7.71971488e-07, 1.30305895e-06, ...,
-4.75553281e-06, -5.09520252e-07, -6.73740855e-06],
[ 1.87471343e-06, 1.10809976e-06, 1.42277977e-06, ...,
-2.39445056e-06, 2.08157541e-06, -3.53570199e-06],
[ 3.62235228e-06, 7.41319898e-07, -3.49176708e-07, ...,
-2.82527907e-06, -8.71546782e-07, -3.88825269e-06],
...,
[ 4.11020044e-07, 2.53134557e-06, 2.02588775e-06, ...,
1.63371394e-06, 1.75639879e-06, -3.22866969e-07],
[-8.12184975e-08, 1.19783257e-08, 1.19906259e-06, ...,
1.12612937e-06, -2.73425855e-06, -1.50807251e-08],
[-1.48969900e-07, 1.02379750e-06, 1.90242890e-07, ...,
-9.31770169e-07, 3.19786912e-07, -1.57491654e-06]],
[[-5.70450429e-07, 2.12182688e-06, -5.69412054e-07, ...,
2.37708527e-06, 9.37329957e-07, 2.78432254e-06],
[-7.37623648e-07, 2.72325232e-06, -1.96874417e-07, ...,
2.58707902e-06, -8.01410389e-08, 2.32243357e-06],
[-1.58156464e-06, 2.49840962e-06, -9.17596458e-07, ...,
1.39761570e-07, -1.76330512e-07, 5.79787961e-07],
...,
[-2.38621266e-06, -8.99388305e-07, -2.34086858e-06, ...,
4.76447796e-06, 2.63580832e-06, 4.52300984e-06],
[-1.04528612e-06, -4.22921727e-07, -1.40656041e-06, ...,
4.72477781e-06, -1.05594628e-06, 4.79468728e-06],
[-1.39863144e-06, -3.56393153e-07, -1.70212073e-06, ...,
3.42515240e-06, -1.34744634e-06, 4.70863707e-06]],
...,
[[-1.86308732e-06, 3.40093890e-07, 4.05648389e-07, ...,
-2.85624522e-06, 8.61272186e-08, -5.85148186e-06],
[-1.24991478e-06, 1.10403278e-06, 2.11822745e-06, ...,
-6.68324617e-08, 1.03962727e-06, -2.65518697e-06],
[-6.01251125e-07, -1.02557637e-06, -9.88862751e-07, ...,
1.19219068e-06, 4.49834173e-06, -1.45729409e-06],
...,
[-2.60819856e-06, -1.14139878e-06, -2.32209565e-07, ...,
2.28403104e-06, 7.43478658e-06, 6.45469349e-07],
[-1.77894845e-06, -3.29765670e-06, -1.96107364e-07, ...,
3.17559439e-06, 8.82660892e-07, 1.14646057e-06],
[-4.46974178e-07, -1.49241340e-06, -9.91849226e-07, ...,
1.50548785e-06, 4.02040655e-06, -7.80036576e-07]],
[[-2.58927496e-06, -6.42217652e-07, -6.51667051e-07, ...,
1.39914167e-06, 1.07433007e-06, 4.62944481e-07],
[ 8.39045230e-07, 1.35565975e-06, 2.19848898e-06, ...,
-1.84449635e-07, 3.03922516e-07, -6.40152619e-07],
[ 2.03991426e-06, 2.96032981e-06, 1.86645511e-06, ...,
1.14277459e-06, -2.05518754e-06, 7.61602543e-07],
...,
[ 1.10087279e-06, 3.53600540e-06, 3.89314588e-06, ...,
-1.54317240e-06, 1.32320950e-06, -2.62412434e-06],
[-4.13975079e-07, 2.61693672e-06, 9.21826295e-07, ...,
4.31760530e-07, 2.19988807e-08, 3.86507114e-07],
[ 5.69702800e-07, 3.26813717e-06, 1.76872363e-06, ...,
-1.34588335e-06, -4.98871043e-07, -2.59585280e-06]],
[[-1.08671813e-06, 1.68348411e-06, 8.88593660e-07, ...,
4.36248556e-06, 1.79194589e-06, 4.90462425e-06],
[-1.70646054e-06, 9.60379945e-07, 4.99692848e-07, ...,
5.04600076e-06, 3.50628649e-06, 7.49790857e-06],
[-5.89765709e-07, 2.06204158e-07, -4.52929175e-07, ...,
4.39870762e-06, 3.38398127e-06, 5.63799640e-06],
...,
[ 1.09576534e-06, 1.18006051e-06, 9.58097062e-07, ...,
2.56989150e-06, 2.55069701e-06, 4.85468318e-06],
[-9.09168160e-07, -1.24791905e-06, -1.05919818e-06, ...,
4.43486532e-06, 4.98945467e-06, 4.77573354e-06],
[-2.84420080e-06, -5.57317398e-07, -1.55925347e-06, ...,
5.27693951e-06, 1.42830967e-06, 4.90967646e-06]]])
… and dimension descriptions:
<Sensor n=65, name='standard_alphabetic'>
UTS(-0.1, 0.005, 140)
Eelbrain functions take advantage of the dimensions descriptions (such as sensor locations), for example for plotting:
p = plot.TopoButterfly(eeg, t=0.130)
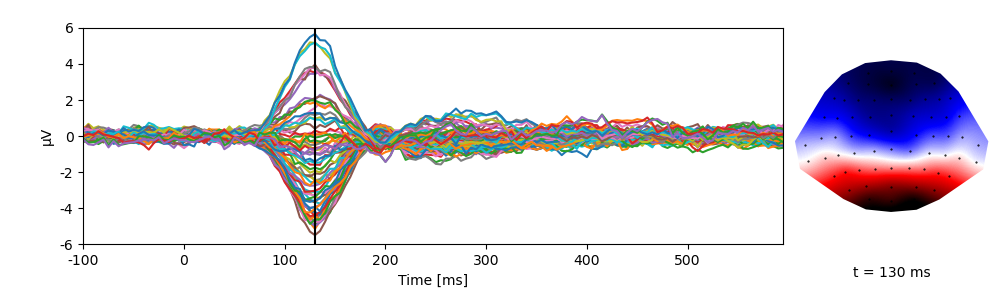
NDVar offer functionality similar to numpy.ndarray, but
take into account the properties of the dimensions. For example, through the
NDVar.sub() method, indexing can be done using meaningful descriptions,
such as indexing a time slice in seconds …
<NDVar 'eeg': 80 case, 65 sensor>
… or extracting data from a specific sensor:
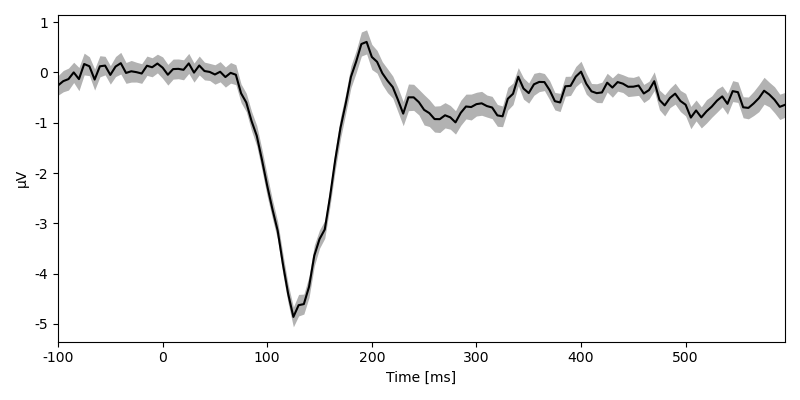
<NDVar 'eeg': 80 case, 140 time>
Other methods allow aggregating data, for example an RMS over sensor …
eeg_rms = eeg.rms('sensor')
plot.UTSStat(eeg_rms)
eeg_rms
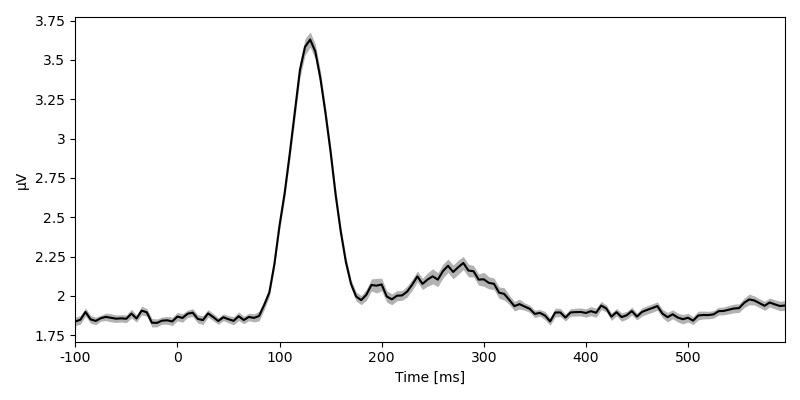
<NDVar 'eeg': 80 case, 140 time>
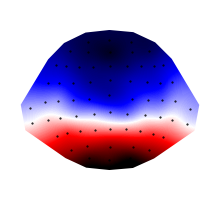
… or a mean in a time window:
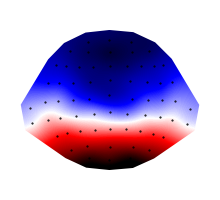
eeg_average = eeg.mean(time=(0.100, 0.150))
p = plot.Topomap(eeg_average)
Dataset
A Dataset is a container for multiple variables
(Factor, Var and NDVar) that describe the same
cases. It can be thought of as a data table with columns corresponding to
different variables and rows to different cases.
Consider the dataset containing the simulated EEG data used above:
Because this can be more output than needed, the
Dataset.head() method only shows the first couple of rows:
This dataset containes severeal univariate columns: cloze, predictability, and n_chars.
The last line also indicates that the dataset contains an NDVar called eeg.
The NDVar is not displayed as column because it contains many values per row.
In the NDVar, the Case dimension corresponds to the row in the dataset
(which here corresponds to simulated trial number):
data['eeg']
<NDVar 'eeg': 80 case, 140 time, 65 sensor>
The type and value range of each entry in the Dataset can be shown using the Dataset.summary() method:
An even shorter summary can be generated by the string representation:
repr(data)
"<Dataset (80 cases) 'eeg':Vnd, 'cloze':V, 'predictability':F, 'n_chars':V>"
Here, 80 cases indicates that the Dataset contains 80 rows. The
subsequent dictionary-like representation shows the keys and the types of the
corresponding values (F: Factor, V: Var, Vnd: NDVar).
Datasets can be indexed with columnn names, …
data['cloze']
Var([0.0261388, 0.995292, 0.930622, 0.824039, 0.170413, 0.18508, 0.233447, 0.807838, 0.249786, 0.267532, 0.193768, 0.131276, 0.115032, 0.124399, 0.947853, 0.17053, 0.848885, 0.204546, 0.214557, 0.89373, 0.234159, 0.859228, 0.239748, 0.823746, 0.825785, 0.920969, 0.191976, 0.934128, 0.138444, 0.156554, 0.81922, 0.933353, 0.967589, 0.127096, 0.822075, 0.995352, 0.863086, 0.872742, 0.043006, 0.832262, 0.23227, 0.850658, 0.289099, 0.820409, 0.997675, 0.964199, 0.00563694, 0.831794, 0.163465, 0.839316, 0.0354823, 0.0793667, 0.277679, 0.871902, 0.827637, 0.939526, 0.856561, 0.261004, 0.283124, 0.164644, 0.841775, 0.136845, 0.00606552, 0.88772, 0.180829, 0.158668, 0.812045, 0.237518, 0.183629, 0.873745, 0.893262, 0.887406, 0.0213108, 0.283401, 0.914039, 0.293586, 0.842077, 0.81942, 0.185291, 0.931266], name='cloze')
… row numbers, …
data[2:5]
… or both, in wich case row comes before column:
data[2:5, 'n_chars']
Var([3, 7, 4], name='n_chars')
Array-based indexing also allows indexing based on the Dataset’s variables:
data['n_chars'] == 3
array([False, False, True, False, False, False, False, True, False,
False, False, False, True, False, False, False, False, True,
False, False, False, False, False, False, True, False, False,
False, True, False, True, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, True, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
True, True, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False])
Dataset.eval() allows evaluatuing code strings in the namespace
defined by the dataset, which means that dataset variables can be invoked
with just their name:
data.eval("predictability == 'high'")
array([False, True, True, True, False, False, False, True, False,
False, False, False, False, False, True, False, True, False,
False, True, False, True, False, True, True, True, False,
True, False, False, True, True, True, False, True, True,
True, True, False, True, False, True, False, True, True,
True, False, True, False, True, False, False, False, True,
True, True, True, False, False, False, True, False, False,
True, False, False, True, False, False, True, True, True,
False, False, True, False, True, True, False, True])
Many dataset methods allow using code strings as shortcuts for expressions involving dataset variables, for example indexing:
data.sub("predictability == 'high'").head()
Columns in the Dataset can be used to define models, for statistics,
aggregating and plotting.
Any string specified as argument in those functions will be evaluated in the
dataset, thuse, because we can use:
data.eval("eeg.sub(sensor='Cz')")
<NDVar 'eeg': 80 case, 140 time>
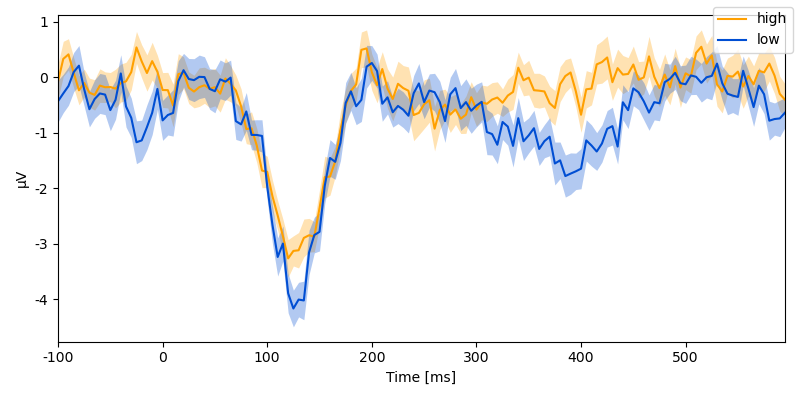
… we can quickly plot the time course of a sensor by condition:
p = plot.UTSStat("eeg.sub(sensor='Cz')", "predictability", data=data)
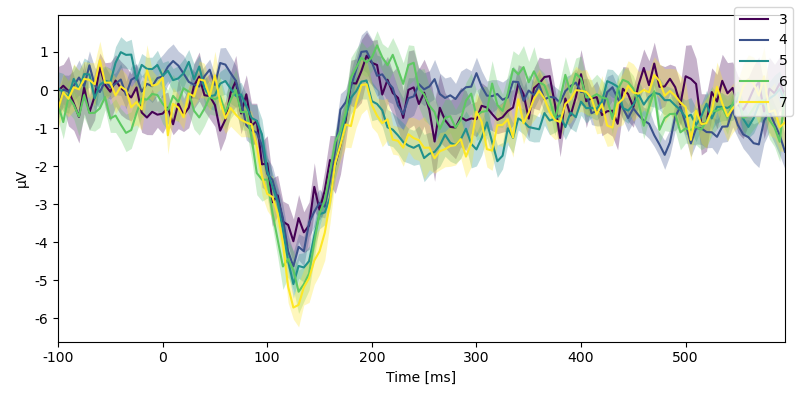
p = plot.UTSStat("eeg.sub(sensor='Fz')", "n_chars", data=data, colors='viridis')
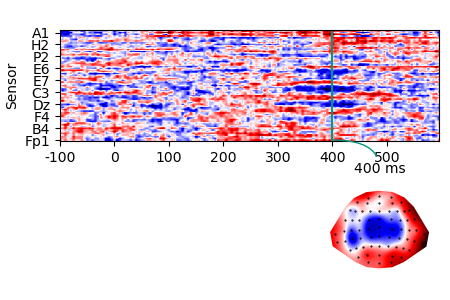
Or calculate a difference wave:
data_average = data.aggregate('predictability')
data_average
difference = data_average[1, 'eeg'] - data_average[0, 'eeg']
p = plot.TopoArray(difference, t=[None, None, 0.400])
For examples of how to construct datasets from scratch see Dataset basics.